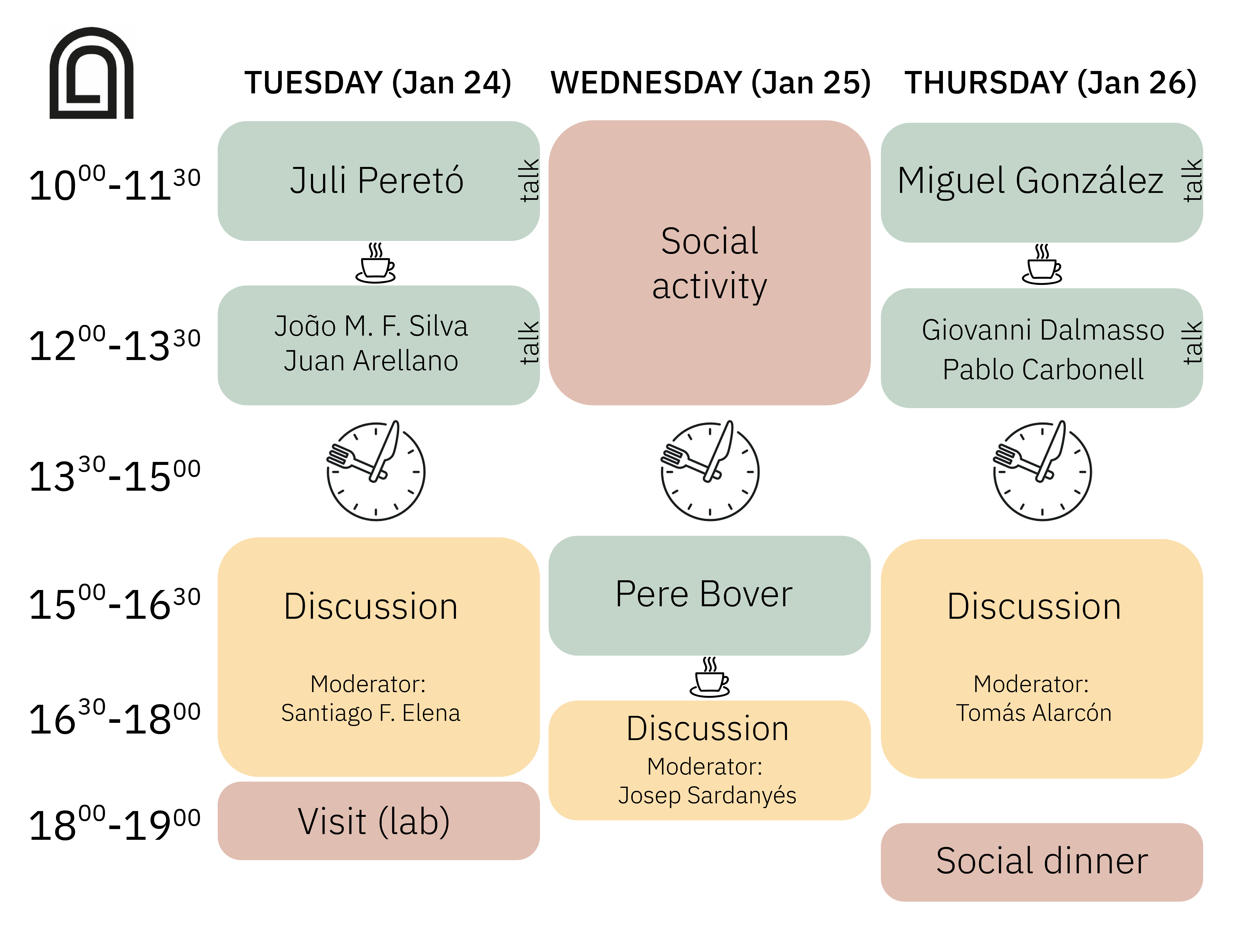
The LSC-Retreat is an initiative born from the collaboration among Laboratorio Subterráneo de Canfranc (LSC),
Centre de Recerca Matemàtica (CRM), and Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV.
This year, the 4th edition of the LSC-Retreat will take place at the Laboratorio Subterráneo de Canfranc (LSC),
Canfranc, Spain, from 24 to 26 January 2023. The aim of this meeting is to encourage scientific discussion
about trending topics on mathematical modelling on biology, ecology, and life systems.
The LSC-retreat 4.0 Organising Committee:
Daria Stepanova (Laboratorio Subterráneo de Canfranc)
J. Tomás Lázaro (Universitat Politècnica de Catalunya)
For more information about the LSC-retreat, please, contact us by e-mail to dstepanova@lsc-canfranc.es.
Conferenciantes Invitados
| Senior Talks: |
|---|
| Pere Bover, Instituto Universitario de Investigación en Ciencias Ambientales (IUCA), Zaragoza, Spain
“DNA damage in paleogenomics: a ‘friendly’ enemy”
Paleogenomics is a relatively new discipline that in the last couple of decades has massively contributed to the understanding of, among others, the taxonomy, phylogeny, phylogeography and ecology of extinct species as well as of ancient populations of extant species. Whereas the first studies from the 1980s were based on relatively short sequences, the improvement in sequencing (Next Generation Sequencing, NGS), wet laboratory protocols and bioinformatic methods during the last decade has allowed us to obtain and analyse a huge amount of data. Although in the pre-NGS studies, the postmortem DNA damage/degradation was already identified as a potential issue in sequence quality, its role in the analyses currently performed is very important to assess the preservation and age of the samples, and to identify possible modern DNA contamination. |
| Miguel González Blanco, Center for Research in Molecular Medicine and Chronic Diseases (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain
“Overview of DNA repair mechanisms in eukaryotes”
The preservation of genome stability is one of the fundamental tasks that a cell must accomplish to faithfully transmit its genetic information to the next generation. However, the integrity of our DNA is under constant threat from multiple sources of damage, both endogenous (oxidative reactions, spontaneous hydrolysis, chemical modifications, replicative stress, etc.) and environmental (ionising radiation, UV-light, reactive compounds, chemotherapeutic drugs, etc.). All these genotoxic agents inflict a myriad of lesions in the DNA of each cell every day. In this talk, I will summarize the repair pathways that our cells employ to counteract each specific type of DNA damage and, therefore, avoid the accumulation of mutations that cause various diseases, including cancer. |
| Pablo Carbonell, Universitat Politècnica de València (UPV), València, Spain
“The extended metabolic space: from evolution to design”
Enzyme promiscuity has often been identified as a latent ability in metabolic networks to evolve novel activities and adapt to new environments. Understanding the mechanisms of enzyme promiscuity, thus, can bring new insights into how pathways have evolved and guide metabolic engineering and directed evolution strategies. To that end, predicting enzyme promiscuity might be attempted solely based on sequence and structural analyses but, more interestingly, they can be combined with the analysis of chemical signatures of biochemical transformations into a multidimensional chemical and sequence-based tree of life. Here, we will use the reaction chemical signatures RetroRules system to model two possible mechanisms for enzyme evolution, the progressive and the regressive models, correlating the results of their simulations to the promiscuity distribution as observed through phylogenetic studies of modern metabolic networks. Based on those models, we will develop an approach to score engineered heterologous pathways in chassis organisms, as predicted by RetroPath, based on the likelihood of the efficiency of the reactions, and will discuss a more general approach based on Selenzyme to prioritize enzyme selection for a given reaction rule and biosensor design by SensBio. Finally, multiobjective optimization strategies for sampling solutions in the extended design space of production- sensing-regulation metabolic circuits will be discussed. References: Carbonell, P., Wong, J., Swainston, N., Takano, E., Turner, N.J., Scrutton, N.S., Kell, D.B., Breitling, R., Faulon, J.L. Selenzyme: Enzyme selection tool for pathway design, Bioinformatics, 34, 2153– 2154, 2018. doi:10.1093/bioinformatics/bty065 Delépine, B., Duigou, T., Carbonell, P., Faulon, J.L. RetroPath2.0: A retrosynthesis workflow for metabolic engineers, Metabolic Engineering, 45, 158–170, 2018. doi:10.1016/ j.ymben.2017.12.002 Carbonell, P., Lecointre, G., Faulon, J.L. Origins of specificity and promiscuity in metabolic networks. Journal of Biological Chemistry, 286:43994-44004, 2011. doi:10.1074/jbc.M111.274050 |
| Juli Peretó, Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV, València, Spain
“In praise of imperfection: tinkering and opportunism in the evolution of metabolism”
François Jacob published an epoch-making article in Science in 1977. Contrary to what incipient molecular biology might indicate, that cells are complex systems with a mesh of highly sophisticated mechanisms perfectly adapted to their function, Jacob invites us to appreciate evolution as something far removed from perfection. Natural selection is more like a tinkerer than an engineer following a set plan and design. Admittedly, the idea of evolutionary bricolage is not original to Jacob but to Darwin himself, who had already formulated it in his study of the co- evolution of flowers and pollinating insects. Jacob’s general formulation of molecular bricolage has materialized in numerous examples and mechanisms of metabolic evolution. A particularly interesting case is that of catalytic promiscuity and its role in enzymatic adaptation and evolution. Today, the precision and specificity observed in many enzymes are confronted with an idea of disordered biology in which noise, infidelity, stochasticity or heterogeneity may also play a crucial role in evolutionary innovation. The exploration of what is chemically possible during oxygen accumulation on Earth is a good example of the role of enzyme plasticity in metabolic evolution. |
| Young researcher talks: |
|---|
| Juan Arellano Tintó, Centre de Recerca Matemàtica (CRM), Barcelona, Spain
“Can mechanical cues guide anastomosis?”
Angiogenesis is the generation of new blood vessels from the pre-existing vasculature. It is a complex, multi-scale process involving cell migration through a heterogeneous and dynamic medium (extracellular matrix, ECM) in response to signaling cues (known as angiogenic factors, AF). In this thesis, we address a process crucial for the formation of a functional vascular network, anastomosis, whereby two growing sprouts join to form a perfused vessel. We will explore the feasibility of a recently proposed hypothesis that mechanical cues guide anastomosis. When endothelial cells (ECs) migrate through the fibrous ECM, focal adhesions form and detach leading to the generation of active traction forces. This results in local deformation and re- orientation of the ECM fibers, creating stress fields that other ECs can sense at a distance. In this thesis, we aim to formulate a proof-of-concept model to ascertain whether these mechanical cues are sufficient to guide anastomosis in a 3D environment. |
| Giovanni Dalmasso, Centre de Recerca Matemàtica (CRM), Barcelona, Spain
“Mathematical modelling of vasculature regression during cartilage condensation”
The growth of an embryo, after it reaches a size of ca. 2mm, heavily depends on a functional vascular system. Unlike other organs, the vascular system needs to be fully functional and constantly remodelling itself to support all tissues from an early stage in embryo development. A crucial question – still not totally understood – is how the 3D architecture of blood vessel networks is controlled during organogenesis. Specifically, in the case of limb development, vasculature is not only essential to supply nutrients but is also fundamental in limb patterning. It has been shown that blood vessels, after creating a functional network, regress from the region where the formation of cartilage condensation begins, while they are maintained in the other parts. Preliminary literature findings and experiments suggest that the transcription factor Sox9 is involved in the regulation of VEGF expression responsible for this phenomenon. In order to fully understand this phenomenon we implemented a hybrid mathematical model with the aim to (1) simulate the contact interactions between endothelial cells (ECs), (2) reproduce the mechanical interactions between ECs, (3) model the Sox9 (pre)pattern, (4) control the Sox9 pattern and (5) model the influence of mechanics on the Sox9 pattern. Combining an approach at the edge between biology and mathematical modelling we provide a first step in understanding how the vasculature network forms inside the limb and how it orchestrates organogenesis. |
| João Marcos Fagundes Silva, Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV, València, Spain
“Temporal rank dynamics of transcripts in SARS-CoV-2-infected cells”
Taylor’s power law is a ubiquitous statistical law in complex systems that describes the distribution of elements by a power relationship between their mean and variance. Here, we used Taylor’s law coupled with a ranking system to analyze the longitudinal stability of transcripts in cells infected with severe acute respiratory virus 2 (SARS-CoV-2) in single-cell RNA-sequencing (scRNA-seq) data. For all three different cell types analyzed (human bronchial epithelial cells and colon and ileum organoid cells), we found that instability increases at later stages of infection. The top 25% genes with the highest accumulated rank were then grouped into stability quartiles. The quartile of the most stable genes showed the highest similarity across cell types, followed by the quartile with the least stable genes. Genes clusters were built based on their rank dynamics. This analysis suggests that the temporal dynamics of some interferon-respondent genes is cell type- specific. Further analyses are underway to dissect the goodness-of-fit of Taylor’s law to scRNA- seq data, as well as to identify gene modules with abnormal behavior and modules that drive instability in these systems. |
Participantes Invitados
| Nombre | Afiliación |
|---|---|
| Alarcón, Tomás | Institució Catalana de Recerca i Estudis Avançats (ICREA), Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Alonso, David | Centre d’Estudis Avançats de Blanes CSIC (CEAB), Blanes, Spain |
| Arellano Tintó, Juan | Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Bover, Pere | Instituto Universitario de Investigación en Ciencias Ambientales (IUCA), Zaragoza, Spain |
| Buceta, Javier | Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV, València, Spain |
| Capitán, José Ángel | Universidad Politécnica de Madrid (UPM), Madrid, Spain |
| Carbonell, Pablo | Universitat Politècnica de València (UPV), València, Spain |
| Dalmasso, Giovanni | Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Elena, Santiago F. | Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV, València, Spain |
| Fagundes Silva, João Marcos | Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV, València, Spain |
| Fontich, Ernest | Universitat de Barcelona (UB), Barcelona, Spain |
| González Blanco, Miguel | Center for Research in Molecular Medicine and Chronic Diseases (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain |
| Guillamon, Antoni | Universitat Politècnica de Catalunya (UPC), Barcelona, Spain |
| Hernández Antolín, Rebecca | Laboratorio Subterráneo de Canfranc (LSC), Canfranc-Estación, Spain |
| Ivancic, Filip | Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Jorba, Marc | Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Lázaro, J. Tomás | Universitat Politècnica de Catalunya (UPC), Barcelona, Spain Laboratorio Subterráneo de Canfranc (LSC) Canfranc-Estación, Spain |
| Martínez España, Lorena | Universitat de València, València, Spain |
| Oro, Daniel | Centre d’Estudis Avançats de Blanes CSIC (CEAB), Blanes, Spain |
| Oteo, Jose Ángel | Universitat de València (UV), València, Spain |
| Otero-Muras, Irene | BioProcess Engineering Group, IIM-CSIC, València, Spain |
| Pedarra, Stefano | Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Peña Garay, Carlos | Laboratorio Subterráneo de Canfranc (LSC), Canfranc-Estación, Spain |
| Peretó, Juli | Institut de Biologia Integrativa de Sistemes (I2SysBio) CSIC-UV, València, Spain |
| Sardanyés, Josep | Centre de Recerca Matemàtica (CRM), Barcelona, Spain |
| Stepanova, Daria | Laboratorio Subterráneo de Canfranc (LSC), Canfranc-Estación, Spain |
Agenda